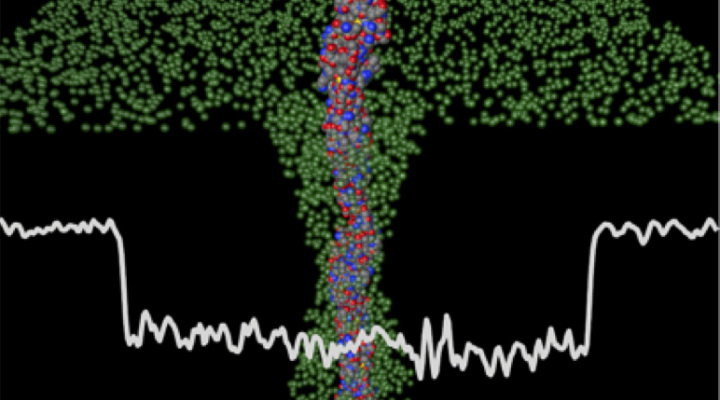
The Open Philanthropy Project recommended a grant of $2,054,142 over three years to the University of Notre Dame to support the development of an instrument that uses a sub-nanometer-diameter pore (i.e. a sub-nanopore) to read the amino acid sequence of whole protein molecules. The collaborative effort led by Dr. Gregory Timp involves researchers at the University of San Diego and Johns Hopkins University. This grant falls within Open Philanthropy's work on scientific research, and was identified through Open Philanthropy's 2016 NIH Transformative Research Award RFP.
Currently, proteomics relies mainly on mass spectrometry (MS) to analyze the structure of proteins. However, MS does not inform on the complete sequence; it lacks sensitivity (it requires about 1 billion molecules); and it is accomplished using an expensive, room-sized apparatus. In contrast, a sub-nanopore, which is about the size of an amino acid residue, reads the primary structure of a single whole protein molecule, although imperfectly, and is embedded in a microfluidic device about the size of a flash-drive.
If it proves out, Open Philanthropy believes this tool could facilitate a wide range of basic biological research and ultimately allow for rapid detection of pathogens, thereby improving the diagnosis and treatment of disease as well as potentially improving our ability to respond to pandemic threats. While the initial applications of this tool are expected in research settings, Open Philanthropy believes it is possible that it could eventually be commercialized and lead to inexpensive methods for protein identification and sequencing in clinical settings.
Using this funding, Dr. Timp and his collaborators will experiment with new sub-nanopore topographies, membrane materials and electrolyte conditions to improve the read fidelity and translocation kinetics, as well as explore new algorithms to discriminate between proteins and identify all twenty proteogenic amino acids and their post-translational modifications.